DeepMind AlphaFold2 & The Future of Biology
DeepMind's AlphaFold2 AI model can predict the three dimensional structure of proteins with an average accuracy of 90%
This article was discussed in our Next Byte podcast.
The full article will continue below.
Protein Structures
A protein’s three dimensional structure determines its functionality but knowing what the structure looks like isn’t easy. Efforts to determine these structures depend on X-ray Crystallography and Cryo-Electron Microscopy (Cryo-EM); both of which offer their own advantages and disadvantages.
Main takeaway, without getting too into the weeds, is that determining these structures manually is a painstaking process and the technology that is currently available limits testability/accuracy of results.
Amino Acids
There are 20 amino acids that act as the building blocks for all proteins and the protein structure is determined by the unique sequence of these amino acids. Christian Boehmer Anfinsen Jr. - 1972 Chemistry Nobel Peace Prize recipient - used this knowledge as the inspiration for his speech where he floated the idea that it should be possible to predict protein shapes based on their amino acid sequences. And thus he kickstarted the ~50 year race to predict protein folds.
The Competition
The Critical Assessment of Structure Prediction (CASP) competition was created by Professor John Moult to accelerate advances regarding protein fold predictions. Participants are given information about a set of proteins and their goal is to come up with a predicted model that is ~90% similar to the structure that has been experimentally determined. Some of the most brilliant scientists from around the world have contributed to this goal via the CASP competition every 2 years since 1994.
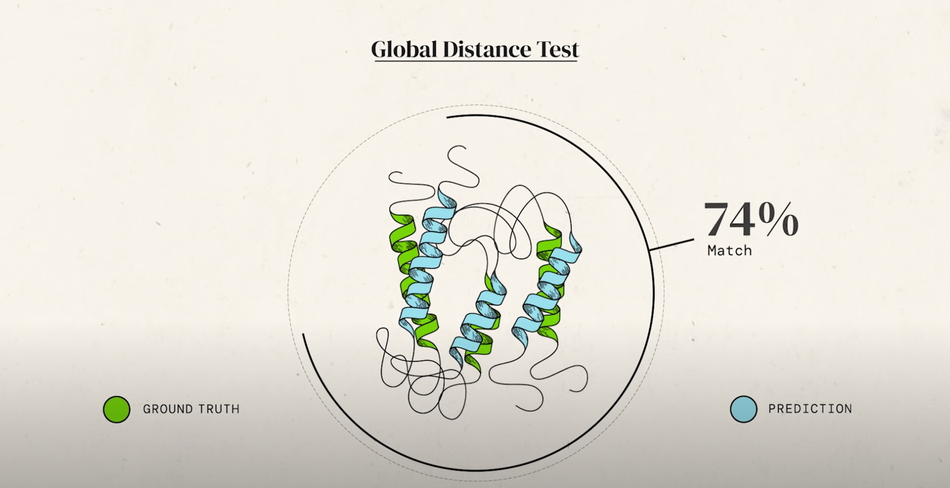 Image 1. Global Distance Test (GDT) used to determine model accuracy
Image 1. Global Distance Test (GDT) used to determine model accuracyHistory of CASP
CASP uses the global distance test (GDT) to determine how similar submitted results are to the experimentally determined protein structures. On a scale of 0 - 100, anything above 90 is recognized as a viable solution. CASP participants had historically struggled to break past a GDT of ~60; however, AlphaFold changed that during CASP 13 in 2018.
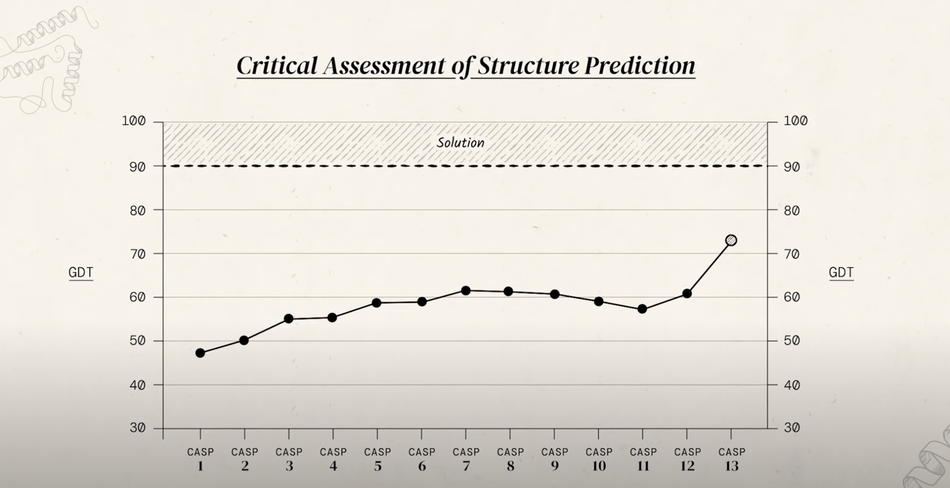 Image 2. Best GDT scores from CASP 1-13
Image 2. Best GDT scores from CASP 1-13The Silver Bullet
AlphaFold is the artificial intelligence (AI) model developed by Alphabet Inc. subsidiary DeepMind, famously known for their AI that beat the world champion Go player, to predict protein folds. Their first iteration of this model predicted the distance between pairs of amino acids by applying deep learning to structural/genetic data and then used this information to come up with a consensus of what the protein should look like. In 2018, this model wowed the scientific community with a GDT of 75.
DeepMind wanted to improve their model and have another shot at CASP; however, they hit a wall using their original approach and had to reassess.
They changed their approach by creating an AI network which utilized geometrical/physical constraints to determine how a protein folds and the model was also given a more difficult task than its predecessor: predict the final structure of the protein instead of the relationship between amino acid pairings.
DeepMind’s hard work paid when the team achieved an average GDT of 90 accuracy for the 7 structures their model had to predict during CASP 14 in 2020. Impeccable timing as well because they predicted the structure of a COVID-19 protein (Orf-8).
Impact
Protein fold predictions will allow researchers to better understand diseases and develop medicine at accelerated rates. Consider Dr. Andrei Lupas from the Max Planck whose team used the AlphaFold algorithm to solve a problem that they've been working on for nearly a decade or how DeepMind was predicting the structures of several under-studied proteins associated with SARS-COV-2 during the peak of the pandemic.
AlphaFold is pivotal addition to scientist's toolboxes for understanding the world and creating a safer future.
